How IT in pharmacogenetics helps to increase drug safety
The European Commission estimates that 197,000 patients die each year from adverse drug reactions. This is partly due to differences in genetic make-up – what works well for Mr Brönnimann misses its target for Mrs Brönnimann or can lead to side effects. This influence can already be examined before the medication is administered, and an application from the Institute for Medical Informatics at the Bern University of Applied Sciences supports this important process Pharmacogenetics (often also called pharmacogenomics/PGx) investigates the connection between a patient’s gene variations and his or her metabolisation of drugs. This knowledge makes it possible to prevent adverse drug reactions (ADRs) in patients with certain gene variations. Currently, the Clinical Pharmacogenetics Implementation Consortium (CPIC) has guidelines for 43 active ingredients [1], and the FDA now lists more than 260 active ingredients with pharmacogenetic biomarkers in the package insert [2]. Both the guidelines and the biomarkers are still rarely taken into account when prescribing and dispensing drugs. Yet ADRs cause avoidable suffering and high costs in the health care system. The EU writes on adverse drug reactions (ADRs):
It is estimated that 197,000 deaths per year in the EU are caused by ADRs and that the total cost to society of ADRs in the EU is €79 billion. Today’s proposals are aimed at further improving the current system. They will save many lives per year across the EU. In addition, they will help to cut red tape by decreasing the administrative burden by ca .€ 145 billion p.a. – European Commission, 2008
[3] In the Swiss health care system, there is a lack of both incentives and efficient and user-friendly IT processes for exploiting the potential of pharmacogenetic markers.
How is pharmacogenetics practised today?
Pharmacogenetics is now an established field and the Pharmacogenomics Knowledge Base (PharmGKB) will soon celebrate its 20th birthday. Pharmacogenetic interaction checks have been available for some time and have become increasingly affordable in recent years. These are offered in Switzerland by some companies and usually support one of two generally describable workflows. In the first workflow, a laboratory sample taken from the patient is sent to the company’s own or a partner laboratory, where the sample is analysed. A report is then generated from the laboratory data and sent either to the patient or to the healthcare provider. The usual way of transport is still by post, although some providers also offer digital dispatch via PDF file. The patients themselves must ensure that their pharmacy and treating physicians receive the pharmacogenetic file. Reports that are difficult for non-specialists to understand sometimes require additional consultation with specialists. Some companies on the market have specialised in producing reports from the laboratory data that are as comprehensible as possible, so that this additional consultation can be saved. In the second workflow, there is a web portal that can be used to create lab orders and retrieve the corresponding analysis reports. Instead of sending a report to the patient or the ordering doctor after the laboratory analysis and the resulting report, the patients receive a digital key with which they can allow their pharmacy or their treating doctors to access the pharmacogenetic data in the web portal. For patients, access to the report is easier to manage than in the first variant.
Why has pharmacogenetics, despite its high relevance and existing providers, not yet made much headway in practice?
One factor for the lack of consideration in the treatment context is the rather small number of studies on WZW evidence (efficacy, appropriateness, cost-effectiveness) in the application of pharmacogenetic guideline information. Therefore, many professional societies still refrain from including pharmacogenetic interaction checks in their own guidelines. However, it can be assumed that this will happen sooner or later. A few pharmacological interaction tests may already be ordered by physicians of all specialities and billed via the health insurance fund. Nevertheless, even these tests are often not used when there is an indication. This suggests that there are further obstacles, for example the expense involved. As indicated above, there is a lack of integrative IT services for the processing and comprehensible presentation of pharmacogenetic knowledge. As a result, an efficient implementation and smooth integration of pharmacogenetic processes into existing processes is often difficult to realise.
A solution for the future
The Institute for Medical Informatics at the Bern University of Applied Sciences has developed an application that seamlessly integrates into the work processes of the involved actors via standardised interfaces (keyword: HL7 FHIR) and takes the diversity of processes into account. The different services of this application can either be used via a web portal or integrated into third-party software, for example, into a practice information system or pharmacy information system. The application knows three process scenarios:
- An initial process when pharmacogenetic data on the patient are not yet available.
- The performance of a pharmacogenetic interaction check if pharmacogenetic data are already available.
- The retrieval of a pharmacogenetic report on the patient.
The initial process concerns the first-time performance of a pharmacogenetic interaction check. Figure 1 shows a generic process, to which variations exist in the application in order to do justice to the different contexts. 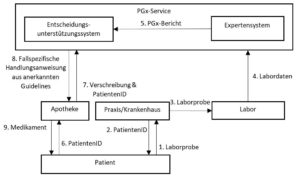
Figure 1: Workflow for a pharmacogenetic interaction check with initial gene analysis.
This process can be illustrated by the example patient Mrs. Brönnimann, who suffers from depression and has been prescribed Escitalopram by her psychiatrist Dr. Baumgartner. To make sure that Ms Brönnimann does not have to wait longer than necessary for convalescence, Dr Baumgartner suggests to Ms Brönnimann that a pharmacogenetic interaction check be carried out.
- Ms Brönnimann agrees and Dr Baumgartner takes a blood sample from her for analysis (1).
- The laboratory order is entered via a web portal. When the order is entered, a patient ID is generated (2).
- The laboratory sample is sent to the laboratory (3),
- analysed there and the laboratory data is transmitted to the PGx service (4).
- An expert system generates a pharmacogenetic report from the laboratory data (pharmacogenetically relevant genotypes of Ms Brönnimann) and transmits it to the decision support system (5).
- Ms Brönnimann is informed that the data have been analysed and that pharmacogenetic interaction checks can be carried out from now on. At the pharmacy, Ms Brönnimann gives the pharmacist the prescription and her patient ID. Before the medicine is dispensed, this data is sent to the decision support system (7).
- With the help of the stored guidelines and the PGx report, the decision support system determines that Ms Brönnimann is a “CYP2C19 Ultrarapid Metabolizer”. This means that Ms Brönnimann metabolises the active substance much more quickly than other people, which in turn means that the concentration of the active substance in the plasma will fall below the desired level much more quickly. Accordingly, there is a high probability that Ms Brönnimann will not respond to the drug [4,5].
- This information is transmitted by the decision support system of the pharmacy software together with some suggestions for alternative drugs whose metabolism is not primarily related to the CYP2C19 gene [8].
- When the pharmacist sees the warning on the display, she picks up the phone and discusses the situation with Dr Baumgartner. They agree that Ms Brönnimann will start the therapy with one of the suggested alternative drugs (9).
Thanks to the pharmacogenetic interaction check, it was thus possible to prevent Ms Brönnimann from taking a drug that is highly unlikely to work for her and therefore potentially only produces side effects. Three years later, the depression half forgotten, Mrs Brönnimann comes to the pharmacy again to order her new cardiovascular drug. Since the necessary data are already available, the decision support system can be triggered automatically before dispensing; it does not detect any increased risk. A classic pharmacogenetic report can also be obtained from the PGx system at any time.
Outlook
In addition to WZW studies on pharmacogenetics, more implementation studies are needed to show how the results of pharmacogenetic research can be made available to the population efficiently. It must be possible for practice/pharmacy/hospital software manufacturers to integrate pharmacogenetic guidelines and the necessary additional processes into their systems with reasonable effort. It should not fail because of IT to treat patients better.
Bibliography
- CPIC Guidelines [Internet]. [cited 11 May 2018].
- Research C for DE and. Table of Pharmacogenomic Biomarkers in Drug Labeling. FDA [Internet]. 27. March 2019 [cited 1 May 2019];
- European Commission – PRESS RELEASES – Press release – Strengthening pharmacovigilance to reduce adverse effects of medicines [Internet]. [cited 30 March 2019].
- Hicks J, Bishop J, Sangkuhl K, Müller D, Ji Y, Leckband S, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clinical Pharmacology & Therapeutics. August 2015;98(2):127-34.
- CPIC® Guideline for Selective Serotonin Reuptake Inhibitors and CYP2D6 and CYP2C19 [Internet]. [cited 25 July 2019].
 Create PDF
Create PDF

 Contributions as RSS
Contributions as RSS Comments as RSS
Comments as RSS
Leave a Reply
Want to join the discussion?Feel free to contribute!